“Haelli restriction,” “enzyme dilution,” “Annaeling,” “Taq polymerase master mix,” “DNA extraction buffer,” “TAS2R38 gene variations” — if none of these terms come up in your daily conversations, you are certainly not alone. But to the students in Valerie Moran’s Biology Extended Learning Opportunity (ELO) last week, these were the concepts and building blocks for an experimental study on genetic predispositions to their individual sensitivities to bitter tastes associated with phenylthiocarbamide (PTC) or glucosinolates.
At the beginning of this day-long, in-school field trip, students were divided into two groups — tasters and non-tasters — depending on whether they found a medical PTC test strip as tasting bitter or neutral. Typically, the ratio of tasters to non-tasters varies between populations but every test group normally has some tasters and some non-tasters. On average, 75% of people can taste PTC while 25% cannot. The students in Moran’s class were evenly divided between tasters and non-tasters (10:10).
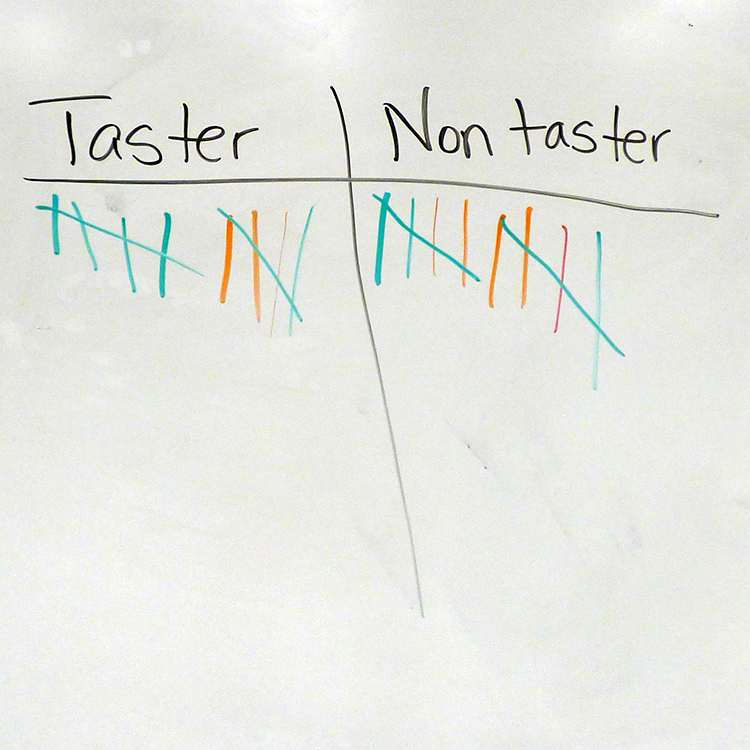
Following this initial assessment of the test groups composition, the scientific research began. Classical Mendelian inheritance dictates who can taste and not taste PTC; however, other factors contribute to the strength of this trait. The combination of inherited alleles along with the interactions between other genes that control facets of taste affect the overall reaction to PTC.
The ability to taste PTC is now linked to the presence of the protein taste receptor 2 member 38 that is encoded by the TAS2R38 gene on chromosome 7. This gene has 2 alleles: the dominant allele (T) confers the ability to taste PTC, while the recessive allele (t) confers the non-taster phenotype.
Sequence analysis of the TAS2R38 gene revealed that PTC taster and non-taster alleles can differ due to SNP’s at 3 distinct locations along the 1143 base pair long coding region of the gene. Each SNP leads to a change in the structure of the receptor protein which affects how well the receptor protein can bind to PTC, which affects the person’s ability to taste PTC.
Students evaluated the presence of one of these SNPs in their own DNA sequence in order to determine their genotype. Students then extracted their own DNA, amplified a segment of the PTC-tasting gene through PCR, used restriction enzymes to identify a possible base pair change in the target sequence, and visualize their genotype on a gel. This allowed students to see how well genotype predicts phenotype.
In doing the extended lab, students learned to explore three molecular biology techniques: DNA extraction, PCR (amplification of their DNA), and gel electrophoresis. The lab also provided students with the opportunity to explore the connection between their genes and their expression, something most people never get a chance to experience.
The type of information that students gathered during the ELO could one day be used to help prevent and treat disease, as well as design new drugs tailored to an individual’s genetic profiles.










